Research
BIDSLab operates at the intersection of data science and biomedicine. Our published and ongoing work research uses image, graph, time series, and genomic datasets to develop AI-based solutions to a range of inverse problems, including image processing and reconstruction, brain network analysis, electrophysiological signal processing for healthcare applications spanning neurology, sleep science, oncology, pulmonology and beyond. Some of our research interests include:
- Regularization strategies for data-limited settings
- Multimodality information integration
- Strategies to address the bias-variance tradeoff in AI
- Geometric learning
- AI-based image enhancement
- AI-based predictive analytics in healthcare
In recent years, our lab has pioneered the use of physics-informed graph neural networks for tau propagation modeling in Alzheimer’s disease, generative adversarial networks for self-supervised super-resolution in positron emission tomography (PET), and sequence-to-sequence architectures for automated sleep staging from wearables.
Graph Theoretic Approaches for Alzheimer’s Progression Modeling
Neurological disorders like Alzheimer’s disease (AD) disrupt both structural and functional connectivity in the brain. At BIDSLab, we have developed techniques based on graph signal processing and geometric learning to interpret and analyze brain connectivity. Our contributions in this domain include graph convolutional neural networks (GCNNs) for staging human subjects on the AD spectrum. Aβ and tau protein aggregates, which are hallmarks of AD, have characteristic spatial patterns with links to disease progression. Graph signal processing approaches are promising for spatiotemporal modeling Aβ and tau. Tau buildup is an important endpoint in ongoing clinical trials for AD. Building on the hypothesis that tau propagates along the structural connectome of the human brain, our recent work uses tau PET and diffusion tensor imaging (DTI) to predict tau spread in the brain. Our contributions in this area include both graph diffusion models and geometric learning models. To address overfitting challenges in low-data settings, we are investigating physics-based regularizers for graph neural networks (GNNs) to enable longitudinal predictive modeling of tau in AD.
Links: NeuroImage 2021 Paper, MICCAI 2020 Paper, IPMI 2019 Paper, ISBI 2019 Paper
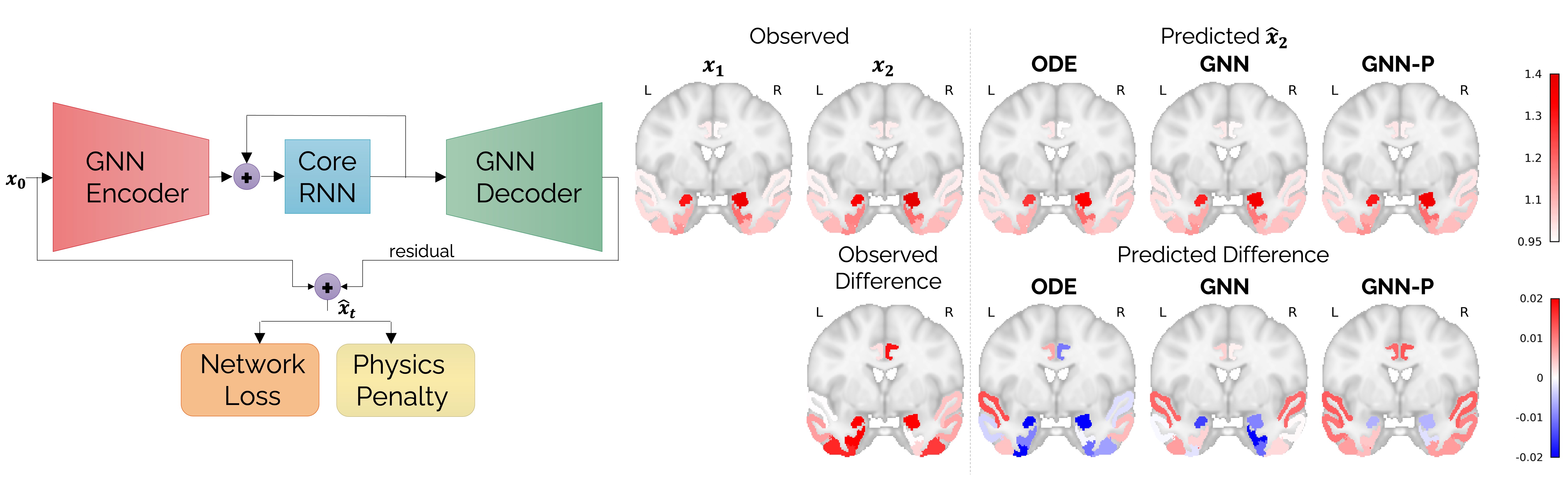
[Left] Schematic showing a physics-informed graph neural network for personalized prediction of longitudinal tau spread. [Right] Brain slices showing the observed distribution of tau at follow-up in a human subject and predictions made from baseline tau PET scans. Results have been compared for a numerical ordinary differential equation (ODE) solver, an unregularized GNN, and a physics-informed GNN (GNN-P).
Super-Resolution Imaging
The quantitative accuracy of PET is degraded by partial volume effects caused by the limited spatial resolution capabilities of PET scanners. We have developed methods for image deblurring and super-resolution for PET that exploits the higher resolution imaging capabilities of high-resolution anatomical magnetic resonance (MR) images. Our contributions to super-resolution PET imaging include supervised models based convolutional neural networks as well as self-supervised models based on generative adversarial networks.
Links: Neural Networks 2020 Paper, IEEE TCI 2020 Paper
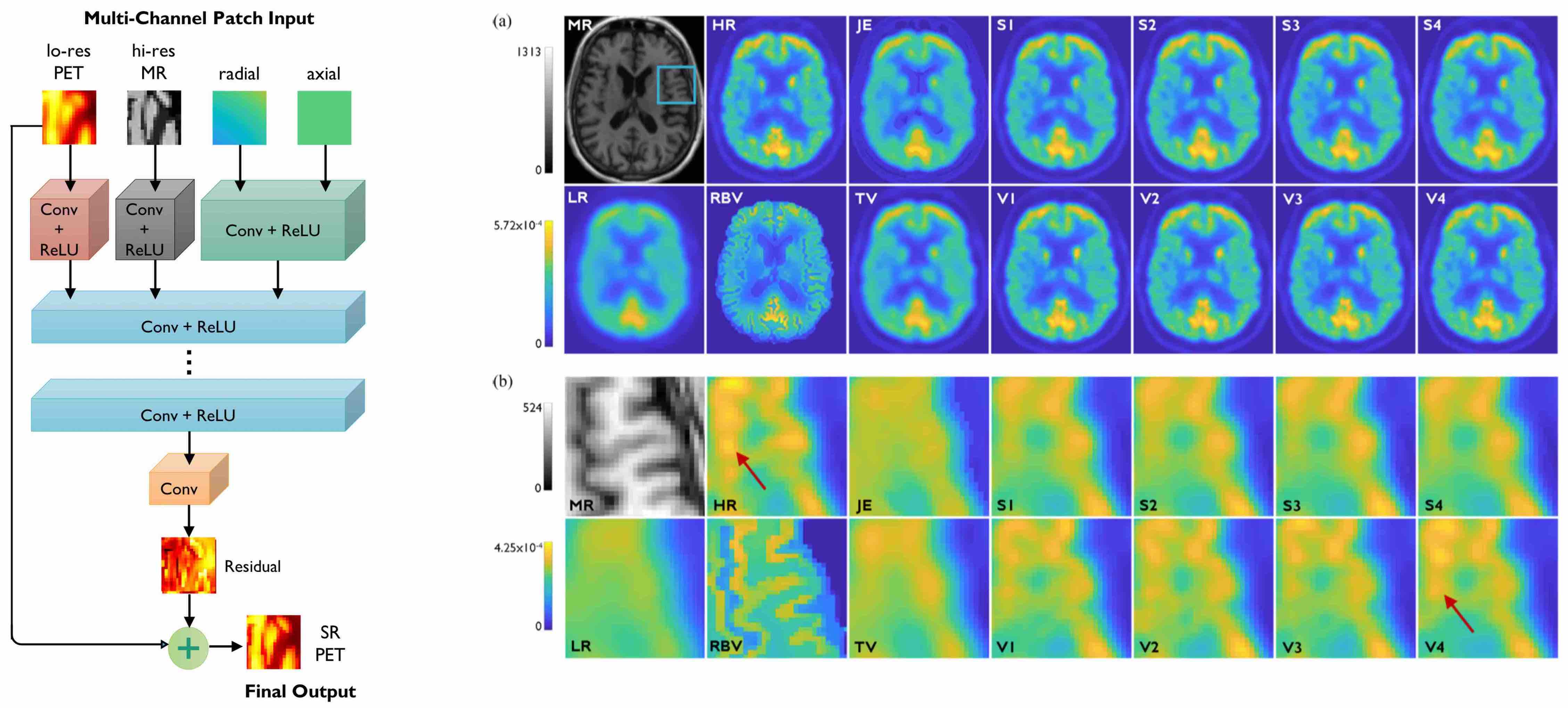
[Left] Very deep CNN based on residual learning with multiple input channels: low-resolution PET, high-resolution MR, and radial and axial location information. [Right] Image slices from human brain scans comparing the performance of the very deep CNN with shallower CNNs and non-deep-learning alternatives.
Image Denoising
The high levels of statistical noise in PET images pose a challenge to accurate quantitation. While deep learning based denoising techniques have been successfully applied to PET imaging, most of these approaches are supervised and require paired noisy and noiseless/low-noise images. This requirement tends to limit the utility of these methods for medical applications as paired training datasets are not always available. Our contributions to PET image denoising include the development of unsupervised PET denoising techniques, e.g., Noise2Void, which do not require paired training data.
Link: PMB 2021 Paper
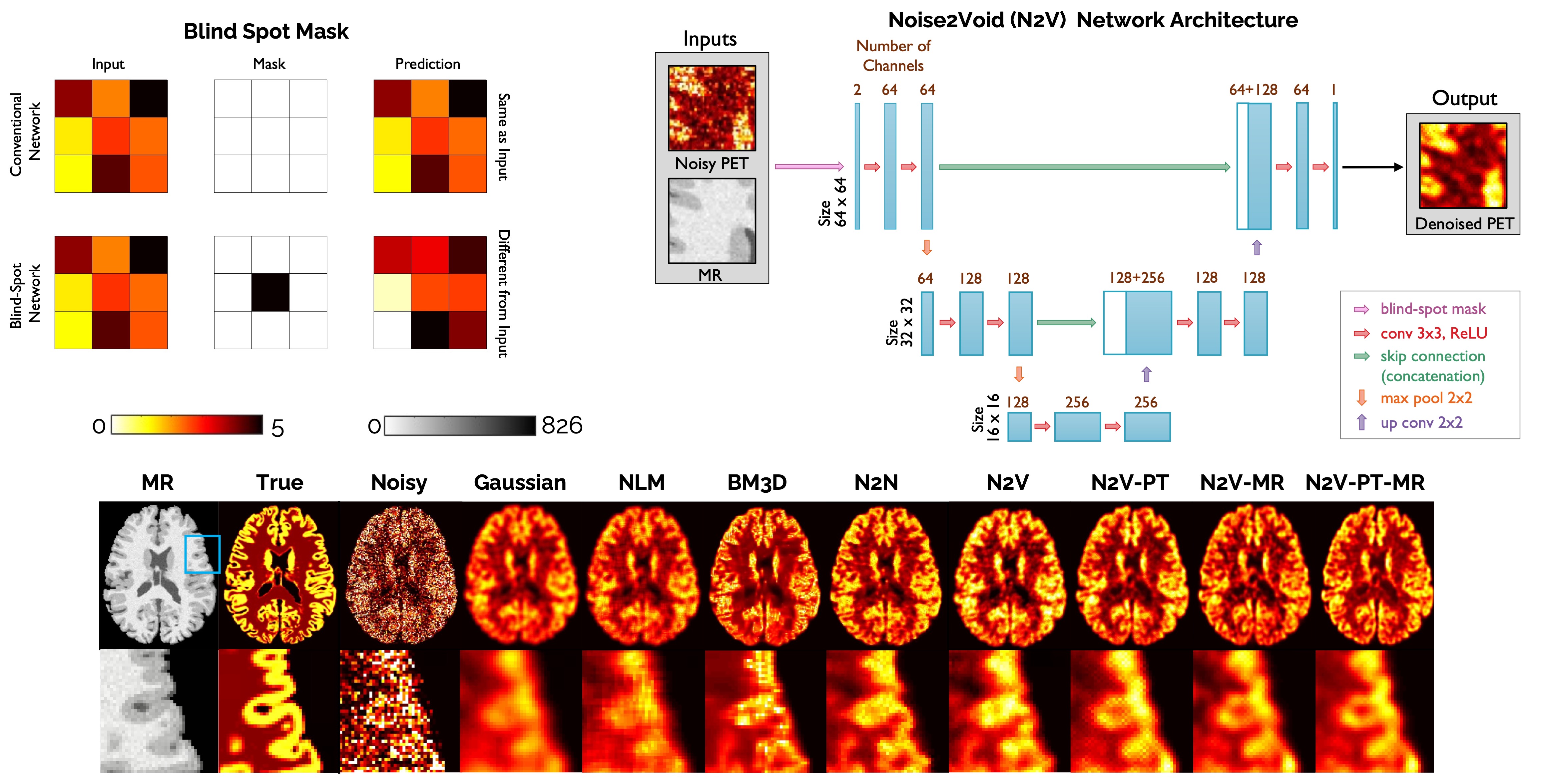
[Top Left] Demonstration of the blind spot masking concept. [Top Right] Network architecture for Noise2Void denoising. [Bottom] Image slices from human brain scans comparing the performance of Noise2Void with alternative denoising techniques.
AI Models for Sleep Time Series
Sleep disturbances, including decline is sleep quality or quantity and microarchitectural disruptions to non-rapid-eye-motion sleep are considered to be among the earliest observable symptoms of several neurological disorders, including Alzheimer’s disease. Monitoring of sleep and activity could thus reveal novel and predictive/early biomarkers for many diseases. We are developing AI models for sleep/activity data based on electroencephalography (EEG), electrocardiography (ECG), and accelerometry. Our research goals include automated multi-class sleep staging and the prediction of a variety of clinical endpoints from time-series datasets. In the specific context of AD, we are investigating potential links between sleep and cognitive decline. We are conducting human sleep studies using consumer-grade wearables that are capable of continuous monitoring of sleep and activity. We have developed AI-based automated sleep staging models based on data from actigraphy devices and smartwatches, such as the Apple Watch.
Link: IEEE TBME 2025 Paper, PLOS One 2023 Paper, bioRxiv Preprint 2022
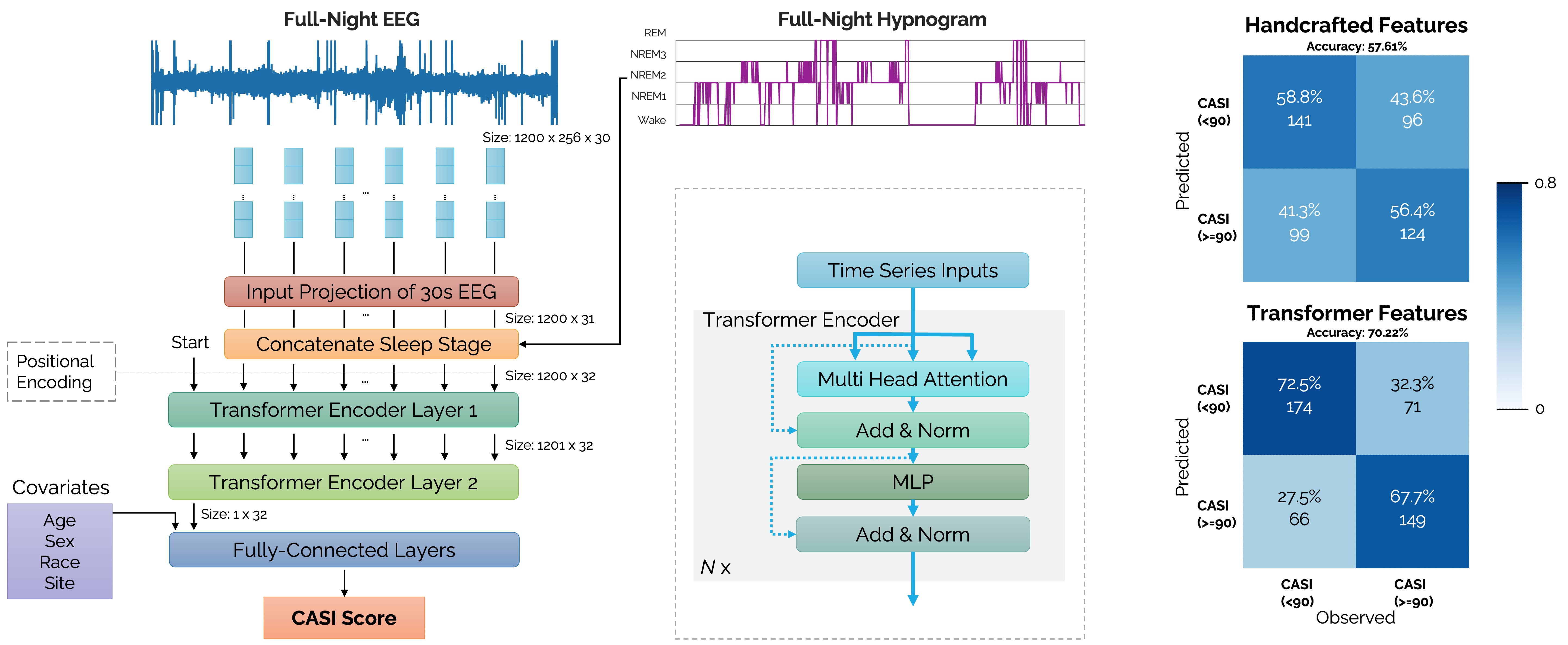
[Left] A transformer encoder network architecture receiving raw EEG time-series inputs concatenated with sleep stage labels (wake, REM, N1, N2, N3) for predicting neuropsychological test scores based on the Cognitive Abilities Screening Instrument (CASI). [Right] Confusion matrices comparing the prediction accuracies of 32 transformer-derived sleep features (overall accuracy 70.22%) with 132 handcrafted sleep features (overall accuracy 57.61%).
Imaging Genetics
Imaging genetics involves the joint analysis of neuroimaging and genetic datasets to assess the impact of genetic variation on brain function and structure. Whereas traditional approaches for imaging genetics have focuses on determining associations between SNPs and imaging phenotypes, we are interested in developing interpretable AI tools that jointly use genomic and radiological inputs to predict clinical endpoints and reveal associations between genotypes and imaging phenotypes that are both region-specific and task-specific.
Link: SNMMI 2021 Abstract
Predictive Analytics
We have a broad interest in developing AI models to predict clinical endpoints from multimodal and heterogenous input types for different clinical problems. Alongside our efforts predicting cognitive decline from sleep data, we are working on a variety of other clinical problems focused on predictive analytics. Examples include predicting cognitive decline from sleep data for Alzheimer’s, therapeutic response from imaging and genetic data for cancer, and exacerbation frequency from clinical and environmental inputs for chronic obstructive pulmonary disease (COPD). These projects rely on large public datasets such as the Alzheimer's Disease Neuroimaging Initiative (ADNI), the COPDGene Study, and The Cancer Imaging Archive (TCIA), the Multi-Ethnic Study of Atherosclerosis (MESA), and biobanks such as the UK Biobank and the MGB Biobank.
Link: IEEE JBHI 2020 Paper
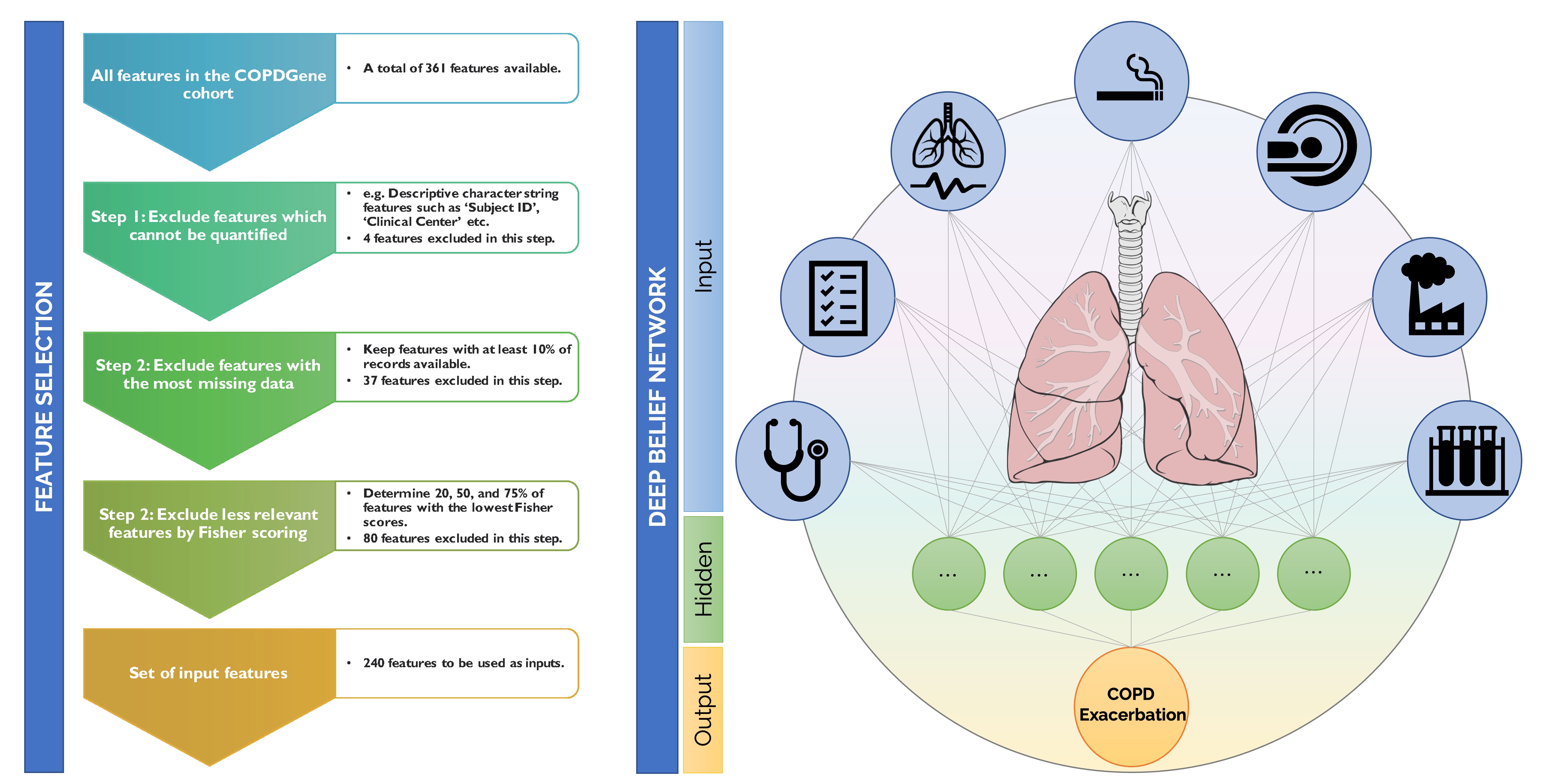
[Left] Pipeline for input feature selection for the COPDGene cohort using Fisher scoring. [Right] Simplified visualization of a deep neural network for predicting COPD exacerbation frequency.